Introducing CellScape™ Whole-Slide Imaging Chamber
Data-driven Assay Expansion: Re-examine Multiplex Immunofluorescence Slides Based on Initial Staining Results
Imagine this: you’ve just completed your multiplex immunofluorescence experiment, and when examining your images, you notice something intriguing or novel about the tissue or a certain subset of cells. In practice, you’d expand the panel to include additional biomarkers and run another experiment; however, the thought of repeating the sample collection, sectioning tissues, and preparing more slides leaves you unmotivated and frustrated that you didn’t include these markers in the first place.
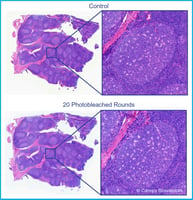
Fortunately, it’s possible to avoid starting from scratch by using high-plex spatial biology technologies like the CellScape™ Whole-Slide Imaging Chamber, which allows you to store and re-stain samples at a later point in time. The CellScape platform allows multiple rounds of staining and imaging to detect dozens of biomarkers from one sample across weeks or months. Because fluorescence signal removal is non-damaging, tissue integrity remains intact during sample storage.
What is data-driven assay expansion?
The case described above is an example of data-driven assay expansion, where initial results from staining and imaging generated new hypotheses and prompted the expansion of the assay to include additional biomarkers. The ability to re-stain your samples over long durations could be especially important when you need more time to analyze data, collect input from collaborators, or wait for new antibodies to arrive.

Data-driven assays expansion using CellScape incorporates rounds of automated staining and imaging, data analysis, and sample storage.
When else would you want to re-examine your samples in spatial biology experiments?
Scenarios like the above aren’t the only times when you might want to re-examine your samples. Additional benefits of data-driven assay expansion include:
- Troubleshooting- If your initial staining didn’t work, you may want to troubleshoot staining conditions or antibody concentrations.
- New antibody- You’ve completed your initial stains, but then a new, key antibody becomes available that wasn’t on hand during the original experiment.
- Quality control- For successful staining, you need properly prepared slides. Before staining your samples with a large, expensive antibody panel, you can test out your slides on a smaller panel of antibodies to eliminate samples that weren’t prepared properly or have degraded.
Advantages of data-driven assay expansion
Data-driven assay expansion allows you to jump right back into data collection with a new lens. This ability has many other advantages in addition to time savings, including:
- Flexibility- The markers you want to analyze don’t have to be all planned out at the start of the experiment. They can be determined part way through staining based on previous results.
- Preserving sample quantity- If your sample is difficult to obtain and/or prepare, or if you just have a limited amount of sample, the ability to re-analyze your slide can facilitate more exploratory experiments without the worry of “wasting” your sample.
- Reduced variability- All markers are assessed in the same tissue sample so this reduces the amount of inter-sample variability that can arise compared to collecting and preparing new samples before staining.
- Error mitigation- The ability to re-stain and analyze your sample means that if you’ve made a mistake during staining, you can repeat the staining process on the same sample. Other spatial biology methods only support one round of data acquisition.
Data-driven assay expansion in action
Data-driven assay expansion is particularly useful in fields like cancer biology and immuno-oncology, where it’s important to localize and monitor protein biomarkers to identify diseased tissue, assess immune responses, and monitor treatment progression. In a recent application note, we demonstrate the benefits of data-driven assay expansion to initially map cell populations before expanding the assay with additional staining and imaging to identify specific tissue structures.
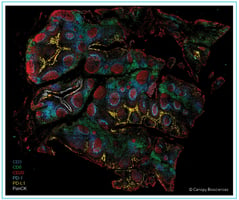
We began with human FFPE lung adenocarcinoma samples and used the VistaPlex Spatial Immune Profiling Assay Kit to identify and locate foundational immune and epithelial cell populations.

Initial assay for immune profiling covered 16 biomarkers.
From there, we identified immune cell markers (e.g., dense CD45 regions as well as high CD20 levels) that suggest the presence of tertiary lymphoid structures (TLS), so we aimed to definitively identify TLS components. TLS are aggregates of immune cells that can be indicators of prognosis in cancer patients1.
We then performed a series of assays every two weeks from the initial stain to probe for tissue architecture features and TLS markers.

Assay expansion of human FFPE lung adenocarcinoma sample in two-week increments.
We confirmed these structures, finding the presence of CD34+ and CD31+ vasculature teeming with CD163+ and CD57+ immune cells as well as markers confirming the presence of dendritic cells, which is a hallmark of TLS. In total, we expanded the assay from the original 16 biomarkers to 40 biomarkers, enabling a detailed understanding of the specific immune system processes in the tumor tissue.

Identification of plasmacytoid dendritic cells characterized as CD3- CD123+ CD4+.
Learn more about data-driven assay expansion in our application note
Re-examine samples up to two years later with CellScape
Our CellScape™ Whole-Slide Imaging Chamber technology uses a safe long-term sample storage buffer to preserve slides after staining and imaging. Since our VistaPlex Assay kits are modular and can be combined with each other or expanded with additional antibodies, use of CellScape with VistaPlex supports assay expansion and provides versatility and simplicity for any spatial biology experiment.
Find out how CellScape can enhance your spatial biology workflow
References
- Chen, Y., Wu, Y., Yan, G., & Zhang, G. (2024). Tertiary lymphoid structures in cancer: maturation and induction. Frontiers in immunology, 15, 1369626. https://doi.org/10.3389/fimmu.2024.1369626