What is Digital Spatial Profiling?
Spatial Biology: What it is, why it matters, and how to access it
Spatial Biology: What it is, why it matters, how to access it.
This article will explore the general topic of spatial biology, providing you with an overview of the field, describing why it is an important area of emerging research, and sharing how you can access spatial biology technologies.
What is Spatial Biology? Spatial Biology Maps Tissue Environments
On a global scale, spatial scientists generate maps for many purposes, from urban planning to monitoring infectious disease outbreaks. Spatial sciences are the studies of the distribution and organization of events and situations in their local environments, including population density, weather patterns, and human activities. These disciplines use visual information (radar, GPS, or satellite imaging for example) to gather data, statistical methods to identify patterns, and even computer modeling to predict future events.
In the same way, but on a smaller scale, spatial biology explores the distribution and organization of patterns occurring in a biological environment, such as a tissue sample. Advanced microscopy and imaging can be used to gather information about a cellular microenvironment based on RNA expression, protein analysis, epigenetics, or the presence of specific mutations in genomic DNA.
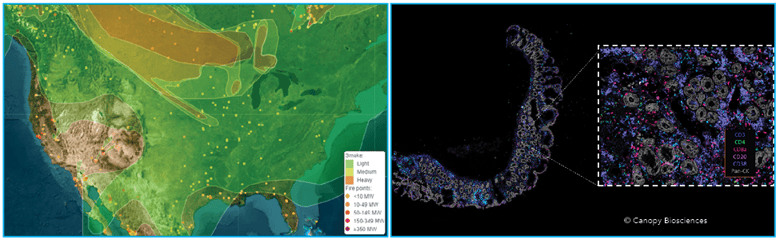
Left: Map of wildfires and smoke in North America (ospo.noaa.gov). Right: Map of immune markers in inflamed colon tissue (Canopy Biosciences).
Using the 2023 North American wildfires as an example, spatial scientists use visual data to create maps that can:
- Identify wildfire locations
- Highlight areas with the densest wildfire smoke
- Predict smoke movement based on wind and weather patterns
- Monitor air quality in individual regions
- Track wildfire changes over time
Just as government spatial science agencies monitor air quality, spatial biologists follow cancer progression and treatment. Microscopic spatial data can be utilized to:
- Identify cancerous tissue
- Highlight regions of immune infiltration
- Predict the most effective therapeutics
- Monitor treatment progress
- Track gene expression changes over time
In addition to oncology applications, spatial biology techniques can also be used to explore organ and tissue development, expand our understanding of degenerative diseases, and design new drugs and therapeutics.
What are Techniques Used for Spatial Biology?
Fluorescence microscopy has been used to explore tissues for decades. Immunohistochemistry (IHC) uses fluorescently labeled antibodies to detect proteins of interest. Fluorescence in situ hybridization (FISH) uses labeled single-stranded DNA oligo probes to highlight specific DNA sequences. An analogous approach, RNA in situ hybridization (RNA ISH), can be utilized to detect RNA, including mRNA, miRNA, and lncRNA. However, these techniques are limited in throughput (only a handful of molecules of interest can be detected at a time) and can be time-consuming and tedious for the bench scientist.
More recent development of higher throughput technologies allowed researchers to explore spatial biology at a much deeper scale. Both high-plex antibody-based imaging and mass spectrometry imaging supported the emergence of spatial proteomics, the profiling of large sets of proteins and their locations within a tissue or cell. By identifying the spatial distribution of proteins within complex tissues, researchers can identify different cell types, learn about protein and cellular interactions, and identify biological responses to treatments.
Spatial barcoding combined with next generation sequencing (NGS) facilitated the emergence of spatial transcriptomics, allowing researchers to sequence RNA and analyze gene expression while retaining information about general tissue regions. The fields of spatial epigenomics and spatial metabolomics explore DNA modifications and small molecules, respectively, in their native locations, with the emergence of traditionally non-spatial technologies paired with spatial outputs to support these newer areas of discovery. Spatial multi-omics is the application of more than one of these omics fields to explore a research question and create powerful datasets to unravel the molecular complexities of biology.
Biology at Single-Cell Resolution
Single-cell analysis allows researchers to isolate and examine individual cells and identify heterogeneity within a tissue or cell population. Traditional high-throughput methods of proteomics, transcriptomics, epigenomics, and metabolomics involve analyzing cell populations in bulk, which masks individual differences that can be essential to understanding biological processes. Single-cell analysis has gained immense attention due to several key, powerful applications:
- Uncovering cellular heterogeneity
- Understanding cellular development and differentiation
- Studying disease mechanisms and immune cell responses
- Biomarker discovery
- Drug development
- Personalized medicine
High-throughput single-cell proteomics has been largely supported by flow cytometry and mass spectrometry. Single-cell RNA-sequencing (scRNA-seq) is a powerful method to provide gene expression data with single-cell resolution and can be achieved via several different workflows, depending on how cells are isolated from each other. Single-cell ATAC-seq, single-cell DNA methylation sequencing, and other emerging technologies support single-cell epigenomics.
Of note, these single-cell analysis techniques require dissociating cells from their tissue environments, a step that loses important information about cell-cell interactions, microenvironment heterogeneity, and localization of protein expression. Making conclusions about cells removed from their microenvironments is akin to drawing conclusions about wildfire smoke from small samples collected from locations without GPS coordinates
ChipCytometry™: Spatial Biology with Single Cell Resolution
Fortunately, advances in imaging and automation technologies have made way for high throughput workflows that allow for detection of many targets while maintaining single cell resolution. The CellScape™ imaging platform from Canopy Biosciences brings the quantitative strengths of spatially-blind techniques like scRNA-seq, flow cytometry, and mass spectrometry to automated high-plex immunofluorescence. With high resolution and high dynamic range imaging, the CellScape combines single-cell resolution, precise protein quantification, and spatial information, providing researchers with a comprehensive spatial biology readout.
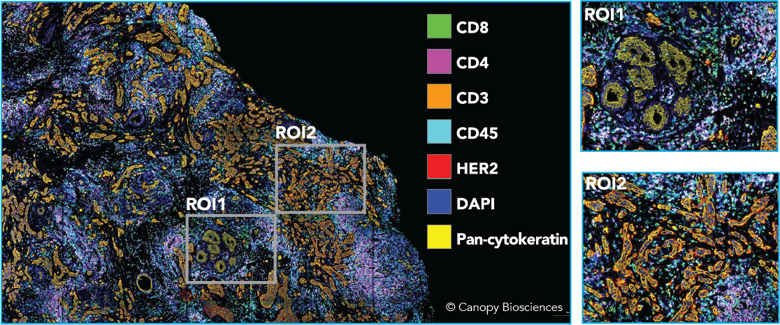
Highly multiplexed image of breast cancer sample. A 21-plex antibody panel was applied to a HER2+ breast carcinoma tissue specimen. Shown above is a subset of biomarkers highlighting tissue architecture and identifying immune cells.
Unique to the CellScape platform, high dynamic range (HDR) imaging enables accurate quantification of the biomarkers being detected. HDR images are created by taking and combining multiple exposures of each fluorescent marker, allowing for detection of both high- and low-expressing targets. After data collection, the images can be analyzed with gating, much like flow cytometry, to classify immune cell types. However, unlike with flow cytometry, cell type data can then be mapped back to their original spatial locations using advanced spatial analysis tools like the Enable Cloud Platform.

Breast cancer tissue with cell types identified. The quantitative biomarker data facilitates cell type and leukocyte type identification, which can be mapped back to the original tissue to generate spatial insights. Shown above, the same HER2+ breast carcinoma sample, with identified cell types mapped back onto the tissue image to allow for additional spatial interrogation.
Spatial relationships can also be explored using advanced analytical techniques, including unsupervised clustering and neighborhood analysis. Investigating the spatial relationships of different types of cells can highlight cell-cell interactions and uncover previously unrecognized biological processes.
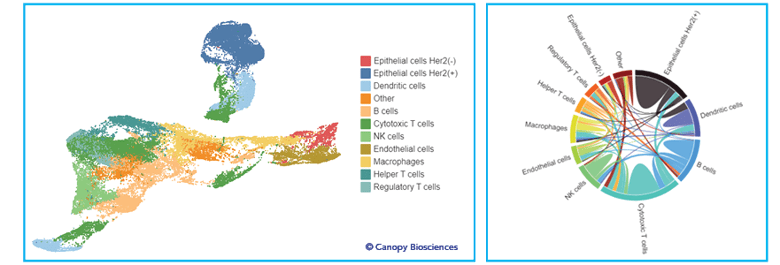
Advanced analysis of spatial biomarker data. The left image shows the result of unsupervised clustering analysis applied to the normalized biomarker expressions of each cell, colored by the identified cell type. The uniform manifold approximation and projection (UMAP) visualization is a way to simplify and visualize multi-dimensional datasets, such as those created by unsupervised clustering. The right plot is a chord diagram generated from the UMAP data; the strengths of cell-cell interactions are represented by the thickness of chords between cell types.
Spatial Proteomics for Validation
Measuring the concentrations of protein biomarkers is key for pharmaceutical development and clinical diagnostics. Because of its relative ease of measurement, RNA is often used as a surrogate for protein expression. RNA can be amplified from small numbers of target molecules and analyzed in a high-throughput manner via next-generation sequencing. However, the relationship between RNA levels and protein quantity is not perfectly linear. There are many post-transcriptional variables that can influence protein levels, including:
- Translation efficiency
- mRNA stability
- Post-translational modifications
- Protein half-life and degradation
- Protein trafficking
Spatial proteomics can be used to complement other cutting-edge technologies provide only information at the RNA level or lack spatial outputs. Instruments like CellScape are opportune as orthogonal validation methods for hypotheses generated from transcriptomic data. As the scientific community raises its demand for rigor and reproducibility, it is also raising the bar for corroborative evidence in biologically relevant contexts to support conclusions.
Spatial and single-cell transcriptomics are appropriate for discovery pipelines, while biomarker characterization, drug development, and ultimately diagnostics and personalized medicine are better supported by more precise spatial proteomics. After all, a spatial scientist would measure the air temperature using a thermometer, not a weather forecast.
How to Access Spatial Biology Technologies
Spatial biology is a revolutionary and exciting field, but it comes with sizable learning and investment curves. Most individual researchers do not have the expertise in the multiple omics techniques and technologies required to carry out a comprehensive spatial biology analysis. Spatial biology projects also require a dedicated instrument or collection of instruments that require capital investments, and these projects are usually most informative and impactful when complementary technologies are paired together to create spatial multi-omic datasets. For these reasons it can be best to partner with an organization with spatial biology expertise like Canopy Biosciences. In addition to providing access to ChipCytometry instrumentation, Canopy Spatial Services™ offers a comprehensive and complementary suite of spatial and single cell services to holistically support spatial multi-omics research questions.



